How do cells achieve the precise synthesis of a complex proteome, given that 98% of the genome is noncoding?
We develop RNA-targeting CRISPR platforms and ASOs that modulate gene expression and reprogram cell fate in disease contexts.

We are developing AI models for predicting RNA 3D structure and cell type/tissue-specific alternative splicing.
1) A web server called kpLogo for position-specific short motif analysis (
Wu & Bartel 2017 Nucleic Acids Research)
2) Network-based prediction of causal genes and pathways in human diseases (Wu et al. 2008 Molecular Systems Biology,
Wu et al. 2009 Bioinformatics)
2026
Peiguo Shi*^, Feiyue Yang*, Tala FNU, Wesley Huang, Alexis O Aparicio, Colin H Kalicki, Aditi Trehan, Michael R Murphy, Esther R Rotlevi, Linqing Xing, Muredach P Reilly, Jianwen Que, Xuebing Wu^
Decoding the MYC locus reveals a druggable ultraconserved RNA element.
bioRxiv, 2026, 2026.01.29.702547; doi: https://doi.org/10.64898/2026.01.29.702547
Michael R Murphy^, Mythily Ganapathi, Esther R Rotlevi, Teresa M Lee, Joshua M Fisher, Megha V Patel, Parul Jayakar, Amanda Buchanan, Alyssa L Rippert, Rebecca C Ahrens-Nicklas, Divya Nair, Shalini S Nayak, Aakanksha Anand, Anju Shukla, Rajesh K Soni, Yue Yin, Feiyue Yang, Enrique J Garcia, Muredach P Reilly, Wendy K Chung, Xuebing Wu^
Pathogenetic mechanisms of muscle-specific ribosomes in dilated cardiomyopathy.
Nature Cardiovascular Research, 2026, 5:51–66
- Research Briefing: Hotspot RPL3L variants drive early onset heart failure through a dual mechanism. Nature Cardiovascular Research (2026)
- See related preprint
2025
Jordan Kesner, Xuebing Wu
Mechanisms suppressing noncoding translation.
Trends in Cell Biology, 2025, 35(7):627-635
Sikandar Azam*, Feiyue Yang*, Xuebing Wu
Finding functional microproteins.
Trends in Genetics, 2025, 41(2):107-118
Tala, Peiguo Shi, Wanwei Zhang, Sanny S.W. Chung, Christopher B Damoci, Yinshan Fang, Qiyue Chen, Anjali Saqi, Yuefeng Huang, Xuebing Wu, Chao Lu, Timothy C. Wang, Jianwen Que
Sympathetic Neurons Promote Small Cell Lung Cancer Through the Beta-2 Adrenergic Receptor.
Cancer Discovery, 2025, 15 (3): 616–632.
2024
Peiguo Shi, Xuebing Wu
Programmable RNA targeting with CRISPR-Cas13.
RNA Biology, 2024, 21:1–9
Imdadul Haq, Jason C. Ngo, Nainika Roy, Richard L. Pan, Nawsheen Nadiya, Rebecca Chiu, Ya Zhang, Masashi Fujita,
Rajesh K. Soni, Xuebing Wu, David A Bennett, Vilas Menon, Marta Olah, Falak Sher
An integrated toolkit for human microglia functional genomics.
Stem Cell Research & Therapy, 2024, 15:104
Jian Ge, Hongxu Ding, Hongxia Shao, Yuefeng Huang, Xuebing Wu, Jie Sun, Jianwen Que
Single cell analysis of lung lymphatic endothelial cells and lymphatic responses during influenza infection.
Journal of Respiratory Biology and Translational Medicine, 2024, 1:10003
2023
Yingyu Zhang, Wanwei Zhang, Jingyao Zhao, Takamasa Ito, Jiacheng Jin, Alexis O. Aparicio, Junsong Zhou, Vincent Guichard, Yinshan Fang, Jianwen Que, Joseph F. Urban Jr, Jacob H. Hanna, Sankar Ghosh, Xuebing Wu, Lei Ding, Uttiya Basu & Yuefeng Huang
m6A RNA modification regulates innate lymphoid cell responses in a lineage-specific manner.
Nature Immunology , 2023, 24:1256–1264
Jordan S Kesner*, Ziheng Chen*, Peiguo Shi, Alexis O Aparicio, Michael R. Murphy, Yang Guo, Aditi Trehan, Jessica E. Lipponen, Yocelyn Recinos, Natura Myeku, Xuebing Wu
Noncoding translation mitigation.
Nature, 2023, 617:395–402
- Spotlight: Mohsen & Slavoff. Noncoding translation: Quality control in the BAG. Molecular Cell . 2023, 83(12):1967-1969
- News Feature: 'Dark proteins' hiding in our cells could hold clues to cancer and other diseases. Nature. 2025, 637(8048):1038-1040
- News release: Illuminating tumor cells with dark proteins
- See related preprint
Peiguo Shi^, Michael R Murphy, Alexis O Aparicio, Jordan S Kesner, Zhou Fang, Ziheng Chen, Aditi Trehan, Yang Guo, Xuebing Wu^
Collateral activity of the CRISPR/RfxCas13d system in human cells.
Communications Biology, 2023 6:334
See related preprint
2022
Jordan S Kesner*, Ziheng Chen*, Alexis A Aparicio, Xuebing Wu
A unified model for the surveillance of translation in diverse noncoding sequences.
bioRxiv, 2022.07.20.500724; doi: https://doi.org/10.1101/2022.07.20.500724
2021
Peiguo Shi^, Michael R Murphy, Alexis O Aparicio, Jordan S Kesner, Zhou Fang, Ziheng Chen, Aditi Trehan, Xuebing Wu^
RNA-guided cell targeting with CRISPR/RfxCas13d collateral activity in human cells.
bioRxiv, 2021, 2021.11.30.470032; doi: https://doi.org/10.1101/2021.11.30.470032
2018
Cheng Zhang, Silvana Konermann, Nicholas J. Brideau, Peter Lotfy, Xuebing Wu, Scott J. Novick, Timothy Strutzenberg, Patrick R. Griffin, Patrick D. Hsu, Dmitry Lyumkis
Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d.
Cell, 2018, 175:212-223
Josh Tycko, Luis A Barrera, Nicholas Huston, Ari E Friedland, Xuebing Wu, Jonathan S Gootenberg, Omar O Abudayyeh, Vic E Myer, Christopher J Wilson, Patrick D Hsu
Pairwise library screen systematically interrogates Staphylococcus aureus Cas9 specificity in human cells.
Nature Communications, 2018, 9:2962
Wei Jiang*, Yuehua Wei*, Yong Long*, Arthur Owen, Bingying Wang, Xuebing Wu, Shuo Luo, Yongjun Dang, Dengke K. Ma
A genetic program mediates cold-warming response and promotes stress-induced phenoptosis in C. elegans
eLife, 2018, 7:e35037
X Shawn Liu, Hao Wu, Marine Krzisch, Xuebing Wu, John Graef, Julien Muffat, Denes Hnisz, Charles H. Li, Bingbing Yuan, Chuanyun Xu, Yun Li, Dan Vershkov, Angela Cacace, Richard A. Young,
Rudolf Jaenisch
Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene
Cell, 2018, 172:979–992
Anthony C. Chiu*, Hiroshi I. Suzuki*, Xuebing Wu, Dig B. Mahat, Andrea J. Kriz, Phillip A. Sharp
Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP
Molecular Cell, 2018, 69:648–663
2017
Xuebing Wu, David P. Bartel
Widespread influence of 3′-end structures on mammalian mRNA processing and stability
Cell, 2017, 169: 905–917
Xuebing Wu^, David P. Bartel^ (^co-corresponding author)
kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences
Nucleic Acids Research, gkx323
http://kplogo.wi.mit.edu
2016
X Shawn Liu*, Hao Wu*, Xiong Ji, Yonatan Stelzer, Xuebing Wu, Szymon Czauderna, Jian Shu, Chikdu S. Shivalila, Daniel Dadon, Richard A. Young, Rudolf Jaenisch
Editing DNA methylation in the mammalian genome
Cell, 2016, 167:233-247
Xiaochang Zhang, Ming Hui Chen, Xuebing Wu, Andrew Kodani, Jean Fan, Ryan Doan, Manabu Ozawa, Jacqueline Ma, Nobuaki Yoshida, Jeremy F Reiter, Douglas L. Black, Peter V Kharchenko, Phillip A. Sharp, Christopher A. Walsh
Cell type-specific alternative splicing governs cell fate in the developing cerebral cortex
Cell, 2016, 166:1147-1162
2015
F. Ann Ran*, Le Cong*, Winston X. Yan*, David A. Scott, Jonathan S. Gootenberg, Andrea J. Kriz, Bernd Zetsche, Ophir Shalem, Xuebing Wu, Kira S. Makarova, Eugene V. Koonin, Phillip A. Sharp, Feng Zhang
In vivo genome editing using Staphylococcus aureus Cas9
Nature, 2015, 520:186-191
2014
Xuebing Wu, Andrea J. Kriz, Phillip A. Sharp
Target specificity of the CRISPR-Cas9 system
Quantitative Biology, 2014, 2:59-70
Xuebing Wu, David A. Scott, Andrea J. Kriz, Anthony C. Chiu, Patrick D. Hsu, Daniel B.
Dadon, Albert W. Cheng, Alexandro E. Trevino, Silvana Konermann, Sidi Chen, Rudolf Jaenisch, Feng Zhang, Phillip A. Sharp
Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells
Nature Biotechnology, 2014, 32:670-676
Sidi Chen, Yuan Xue, Xuebing Wu, Le Cong, Arjun Bhutkar, Eric Bell, Feng Zhang, Robert Langer, Phillip A. Sharp
Global microRNA depletion suppresses tumor angiogenesis
Genes & Development, 2014, 28:1054-1067
2013
Xuebing Wu, Phillip A. Sharp
Divergent transcription: a driving force for new gene origination?
Cell, 2013, 155:990-996
Albert E. Almada*, Xuebing Wu*, Andrea J. Kriz, Christopher B. Burge, Phillip A. Sharp (*equal contribution)
Promoter directionality is controlled by U1 snRNP and polyadenylation signals
Nature, 2013, 499:360–363
Le Cong, F Ann Ran, David Cox, Shuailiang Lin, Robert Barretto, Naomi Habib, Patrick D Hsu, Xuebing Wu, Wenyan Jiang, Luciano A Marraffini, Feng Zhang
Multiplex genome engineering using CRISPR/Cas systems
Science, 2013, 339 (6121):819-823.
Patrick D. Hsu, David A. Scott, Joshua A. Weinstein, F. Ann Ran, Silvana Konermann, Vineeta Agarwala, Yinqing Li, Eli J. Fine, Xuebing Wu, Ophir Shalem, Thomas J. Cradick, Luciano A. Marraffini, Gang Bao, and Feng Zhang
DNA Targeting Specificity of the RNA-guided Cas9 Nuclease
Nature Biotechnology, 2013, 31:827-832
Anna Lyubimova, Shalev Itzkovitz, Jan Philipp Junker, Zi Peng Fan, Xuebing Wu, Alexander van Oudenaarden
Single-molecule mRNA detection and counting in mammalian tissue
Nature Protocols, 2013, 8:1743–1758 (download software)
Yong Chen, Xuebing Wu, Rui Jiang
Integrating human omics data to prioritize candidate genes
BMC Medical Genomics, 2013, 6:57
2010
Xuebing Wu, Shao Li
Cancer gene prediction using a network approach
Chapter 11 in: Cancer Systems Biology (Ed. Edwin Wang). Series: Chapman & Hall/CRC Mathematical & Computational Biology, USA: CRC Press 2010. [Amazon]
2009
Xuebing Wu, Qifang Liu, Rui Jiang
Align human interactome with phenome to identify causative genes and networks underlying disease families
Bioinformatics, 2009, 25 (1): 98-104 [Data]
Wanwan Tang, Xuebing Wu, Rui Jiang, Yanda Li
Epistatic module detection for case-control studies: A Bayesian model with a Gibbs sampling strategy
PLoS Genetics, 2009, (5):e1000464
Rui Jiang, Wanwan Tang, Xuebing Wu, Wenhui Fu
A random forest approach to the detection of epistatic interactions in case-control studies
BMC Bioinformatics, 2009, 10 (Suppl 1):S65
2008
Xuebing Wu, Rui Jiang, Michael Q. Zhang, Shao Li
Network-based global inference of human disease genes
Molecular Systems Biology, 2008, 4:189.
Xuebing's undergraduate publications
Xuebing Wu, Zhirong Sun, Rui Jiang
Logic motif of combinatorial control in transcriptional networks
Nature Precedings, 2008, https://doi.org/10.1038/npre.2008.2227.1
Zhirong Sun, Xuebing Wu
Systems biology: new interdisciplinary science
Communication of China Computer Society, 2006, 2 (3):56-65 (Cover story, review, in Chinese)
Xuebing Wu, Yuanjie Li, Huaiyu Wang
An intelligent transportation system based on virtual force
Physics and Engineering, 2005, 2 (in Chinese)
| 2025/07/03 |  |
 |
Xuebing Wu
|
 |
Michael MurphyAssociate Research Scientist (Senior Postdoc)
|
 |
Peiguo ShiAssociate Research Scientist (Senior Postdoc)
|
 |
Aditi TrehanPhD student, Integrated Program in Cellular, Molecular, and Biomedical Studies, Columbia University
|
Colin Kalicki
PhD student, Systems Biology, Columbia University
|
|
 |
Feiyue Yang
PhD student, Systems Biology, Columbia University
|
 |
Wesley Huang
MD/PhD student, Columbia University
|
 |
Esther R. Rotlevi
PhD student, Neurobiology and Behavior, Columbia University
|
 |
Haikun Wang
M.S. Biotechnology student, Columbia University
|
 |
Mengyuan Chen
MS student, Biostatistics, Columbia University
|
 |
Linqing Xing
Postdoctoral Research Scientist
|
Andrea Mojsoska
Undergraduate student
|
Jordan Kesner 8/1/2020-10/1/2023, PhD student -> Postdoc
Katherine Da 9/1/2023-11/30/2023, Visiting undergraduate student, Dartmouth College
Yue Yin 11/01/2021-07/01/2022, Barnard Summer Research Institute, undergraduate student
Alexis Aparicio 02/19/19-08/19/2022, Technician B
Jonathan Algoo 9/20/21-12/14/2021, Rotation student, integrated program
Zhou "Zanis" Fang 11/01/19-12/06/2021, DSI Scholar -> Postdoc
Ziheng "Calvin" Chen 1/15/19-8/10/2020, Master's student, Columbia BME
Qijin Yin 11/25/19-5/10/2020, Visiting PhD student from Tsinghua University
Ellen Alt 5/28/19-8/1/19, Barnard Summer Research Institute, undergraduate student
|
 |
|
|
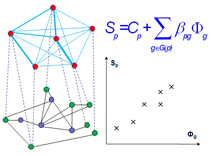
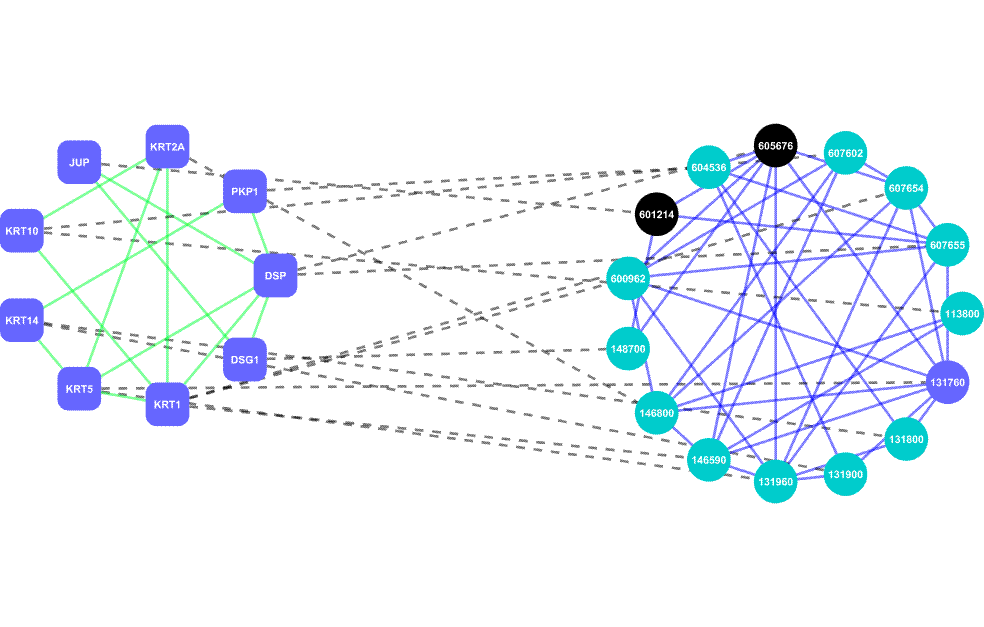
AlignPI is the abbreviation of "Align Phenome & Interactome." We have performed the first alignment of human phenome and interactome network, and identified 39 bi-modules and made predictions of candidate genes for 70 diseases. Bi-module consists of two inter-connected modules: a network of diseases and a network of genes.
Links: The AlignPI website provides a search interface for all identified bi-modules and candidate gene predictions, and corresponding functional enrichment analysis. Citation: Wu X, Liu Q, Jiang R (2009) Align human interactome with phenome to identify causative genes and networks underlying disease families. Bioinformatics, 25 (1): 98-104 |
2/3/26 Xuebing gave an invited seminar at UMass RNA Therapeutics Institute
1/28/26 Peiguo and Feiyue's preprint on MYC saturation mutagenesis available in bioRxiv
1/6/26 Michael's mutant RPL3L ribosome paper has been published in Nature Cardiovascular Research
11/19/25 We receinved funding from the Lewis Katz Cardiovascular Research Prize
10/9/25 Andrea joined the lab as an undergrad researcher
8/22/25 Michael gave a talk on his RPL3L work in CSHL mRNA processing meeting
6/26/25 MD/PhD student Wesley joined the lab
6/2/25 Biostat MS student Mengyuan joined the lab
6/1/25 Linqing joined the lab as a postdoc
5/19/25 Biotech MS student Haikun joined the lab
4/14/25-4/17 Xuebing gave invited seminars at UCLA, UCSF, and Stanford
3/31/25 Esther joined the lab as PhD student #4
1/29/25 Xuebing talked about our work on noncoding translation and microproteins in a Nature news feature
1/15/25 Xingpei Zhang started rotaion in the lab
1/3/25 Michael's work on RPL3L in heart failure is online in bioRxiv
1/2/25 Alex and Feiyue's review on functional microprotein is published in Trends in Genetics
11/8/24 Peiguo's collaborative work with the Que lab is published in Cancer Discovery
10/22/24 Jordan's review on noncoding translation is published in Trends in Cell Biology
8/27/24 Henry Le started rotaion in the lab
8/15/24 Xuebing gave an invited seminar at Tsinghua University, Beijing
7/26/24 Xuebing speaks at the 2024 International Conference on Life Sciences, Guiyang, China
7/18/24 Xuebing is invited to present in 2024 STXBP1 Summit
7/8/24 Colin and Feiyue join the lab as PhD studnet #3 and #4!
7/8/24 Wesley Huang joins the lab for his summer research rotation
6/24/24 Xuebing speaks at the 50th Sharp lab reunion symposium “Half A Century of Sharp Minds”
6/10/24 Caroline Ugoaru joins the lab for her T35 summer research as an MD student
6/7/24 Xuebing speaks at the 2024 High-Risk, High-Reward Research (HRHR) Symposium at Bethesda
5/19/24 Peiguo's Cas13 review was published online
5/13/24 Xuebing was selected as a 2024 Schaefer Research Scholar
4/19/24 Xuebing speaks at the first Healthspan Extension Summit at Columbia
4/3/24 Xuebing is invited to speak at the inaugural RNADay@Penn 2024
3/26/24 We received the MIND Prize to test a new idea for treating neurodegeneration
2/1/24 We received our first R01 grant from NHLBI!
1/8/24 We welcomed rotation students Feiyue Yang (Systems Biology) and Sebastian Ho (Nutritional & Metabolic Bio)
12/20/23 We received a grant from the Million Dollar Bike Ride (MDBR) Grant Program to dissect STXBP1 3’ UTR regulation for therapeutic development
12/19/23 We received a grant from the Ines Mandl Research Foundation (IMRF) to develop an off-the-shelf gene editing therapy for Marfan Syndrome
11/17/23 We received a grant from the Cure Alzheimer’s Fund to study noncoding translation and feedback loops in Alzheimer's disease!
11/10/23 Xuebing is invited to give a seminar at Duke Biology and RNA Center @ Durham
11/9/23 Xuebing is invited to give a seminar at NIEHS @ Durham
10/17/23 Xuebing is invited to give a seminar at Koch Institute/MIT
10/14/23 Xuebing gave a talk at EMBL@Heidelberg, Germany, during the 2023 non-coding genome symposium
10/13/23 Xuebing is invited to give a seminar at Heidelberg University Hospital @ Heidelberg, Germany
10/11/23 Xuebing is invited to give a seminar at Max Planck Institute for Molecular Genetics @ Berlin, Germany
9/20/23 We received the inaugural Glenn Foundation Discovery Award !
9/18/23 Our NCI admin supplement grant with Que lab has been funded!
9/13/23 We welcome visiting student Katherine Da, Biology and CS @ Dartmouth College
9/4/23 We receive a 2nd grant from Longevity Impetus Grants !
8/28/23 We welcome graduate student Colin Kalicki for his 1st rotation
7/31/23 Xuebing gave a talk in the Basic Cardiovascular Sciences (BCVS) conference in Boston
7/3/23 Our collaborative work with Yuefeng Huang lab was published at Nature Immunology
7/1/23 We receive a grant from HICCC/Inter/Intra-Programmatic Pilot Program (IPPP) to target lung cancer using CRISPR/Cas13
5/30-6/4/23 Xuebing, Michael, and Jordan attended 2023 annual RNA meeting at Singapore. Michael won a poster award. Xuebing gave a talk on RPL3L ribosomes.
4/19/23 Xuebing was invited to visit U of Chicago by postdocs at the Department of Human Genetics
4/12/23 Our paper titled noncoding translation mitigation has been published online at Nature. See CUIMC news release Illuminating tumor cells with dark proteins
3/28/23 Our paper on Cas13 collateral activity has been published online at Communications Biology
3/21/23 Jordan has been selected as one of the two recipients of the 2023 Dean’s Award for Excellence in Research! Congratulations, Jordan!
10/14/22 Xuebing and Michael each gave an invited virtual talk to the Toronto RNA Club
9/29/22 Xuebing gave an invited virtual seminar at the University of Texas at Dallas
9/6-9/22 Xuebing and Michael each gave a talk at the 18th translational control conference at CSHL
8/31/22 Xuebing gave an invited talk at the first FASEB conference on RNA and immunity.
8/25/22 Congrats Dr. Jordan Kesner! First PhD from the lab!
7/21/22 Lab releases the 2nd bioRxiv preprint, led by Jordan, Ziheng, and Alex
6/4/22 Jordan gave a talk at RNA society meeting 2022
5/25/22 Congrats to Aditi for passing her qualifying exam!
5/4/22 Xuebing gave an invited talk at Weill Cornell and MSKCC
5/3/22 Xuebing gave an invited virtual talk at Prevail Therapeutics / Eli Lilly and Company
4/18/22 Lab welcomes Yang Guo as a postdoctoral research scientist !
11/30/21 First preprint from the lab, led by Peiguo: RNA-guided cell targeting with CRISPR/RfxCas13d collateral activity in human cells
11/16/21 Lab welcomes Barnard/Columbia undergraduate student Yue Yin!
10/14/21 Lab receives Longevity Impetus Grants to study aging!
9/20/21 Lab welcomes Jonathan Algoo as a rotation student in the Fall!
6/21/21 Lab receives 2021 Paul A. Marks Scholars Award , which provides unrestricted fund for 3 years
6/3/21 Lab receives Columbia Precision Medicine Pilot Grant , which supports our study on specialized ribosomes in the heart
5/27/21 Aditi Trehan became PhD student #2 of the Wu lab!
5/18/21 Lab receives the 2021 Pershing Square Sohn Prize for Young Investigators in Cancer Research, which supports our study on cancer-targeting using CRISPR/Cas13 and machine learning. Also see CUIMC News
4/12/21 Aditi Trehan started her rotation in the lab!
10/6/20 Lab receives NIH Director's New Innovator Award (DP2) for genome-scale study of mRNA noncoding functions using CRISPR/Cas13. Also see CUIMC News
9/1/20 Zanis joined the lab as a Postdoc!
8/3/20 The Wu lab welcomes our first PhD student Jordan Kesner!
6/15/20 Xuebing is named a 2020 Pew-Stewart Scholar for Cancer Research, which will support our study of how cancer cells reprogram mRNA structures on a genome-scale. Also see Columbia HICCC News
4/1/20 Lab's COVID-19 research highlighted in HICCC news article Columbia University Cancer Researchers in the Fight Against COVID-19
2/21/20 Xuebing's faculty profile/interview article at Dept. Systems Biology and Cancer Center / HICCC
2/10/20 Shruti Verma joined the lab from Columbia Engineering
12/17/19 Xuebing is named Highly Cited Researcher in 2019 by Web of Science
12/03/19 Michael's AHA postdoc fellowship application was funded. Congratulations!
11/25/19 Qijin Yin started as a visiting scholar. Welcome Qijin!
11/01/19 Welcome four new members to the lab: Peiguo Shi (postdoc), Zanis Fang (MS/DSI Scholar), Katherine Liu and Yixuan Li (undergrad)
10/24/19 Lab moves to our permanent space in P&S
06/28/19 Xuebing gave a talk at the RNA therapeutics conference at UMass Med School.
06/14/19 Xuebing gave a video-taped talk at the RNA Society Annual Meeting in Krakow, Poland.
05/28/19 Ellen Alt joined the lab as a summer research student. Welcome Ellen!
03/01/19 Michael Murphy joined the lab as a staff associate. Welcome Michael!
02/19/19 Alexis Aparicio joined the lab as our first Technician. Welcome Alex!
01/15/19 Ziheng (Calvin) Chen, a graduate student in the BME Master of Science program joined the lab. Welcome Calvin!
12/07/18 Xuebing won the The RNA Society/Scaringe Young Scientist Award 2019
11/01/18 Lab officially started!
We are seeking passionate researchers of all stages (postdocs, graduate students, undergraduates, technicians, and high school students) to join our team.
Graduate student looking for rotation or postdoc opportunity: please email me (xuebing.wu@columbia.edu) directly.
Graduate programs we are affiliated with: Integrated Program in Cellular, Molecular, and Biomedical Studies (CMBS); Systems Biology (also a track in the CMBS program); Neurobiology and behavior; Nutritional and Metabolic Biology.
Undergraduate students: If you are interested in becoming a graduate student in the lab, you need to first get admitted to one of the PhD programs at Columbia. Once you are admitted, you can choose to rotate or join the lab. We also welcome passionate and creative undergraduate students who can work in the lab full-time during summer or as a visiting student during regular semester. Dr. Xuebing Wu is a mentor of the Summer Undergraduate Research Fellowship (SURF) program and the Barnard Summer Research Institute.